Cancer Research



Overcoming Challenges in Cancer Genomics
Cancer genomes are notoriously complex—with abnormal chromosome structures, somatic variations, and intricate rearrangements caused by events such as chromothripsis. These obstacles can make the critical, accurate detection of SVs with traditional genomic tools a daunting task. They also create significant obstacles in mapping tumor progression, which is critical for understanding cancer development and treatment resistance.
No single technology is capable of interrogating the complexities of cancer genomics:
- Short-read sequencing: Meant to detect SNPs and indels, and as a result, may miss SVs and copy number variants (CNVs) in key cancer genes and highly repetitive regions
- Long-read sequencing: Expensive and subject to sequencing artifacts, with limited detection of large-scale events
- Karyotyping: Limited to very large-scale events
- Fluorescence in situ hybridization: Targeted and low resolution, lacks a whole-genome view of structural variation
- Chromosomal microarray analysis: Limited resolution and detection of balanced translocations and inversions
EGM using the OhmX Platform provides a novel view of the genome. The OhmX Platform empowers researchers with a full picture of genomic variation that includes critical and often undetected SVs in both somatic and germline samples. The results can include anything from deletions and duplications to complex aberrations that will help unravel the complexity of a unique cancer genomic profile.
Why Choose the OhmX Platform for Your Cancer Research?
Cancer-Relevant SV Detection
Many cancers are driven by genomic rearrangements—including deletions, duplications, inversions, and translocations. These alterations can disrupt tumor suppressors, create oncogenic fusions, or drive genome instability, yet they are often missed or poorly resolved by standard sequencing and cytogenetic techniques. EGM delivers high-resolution, genome-wide SV detection, capturing both known and novel rearrangements that are critical for cancer discovery, stratification, and therapeutic development.
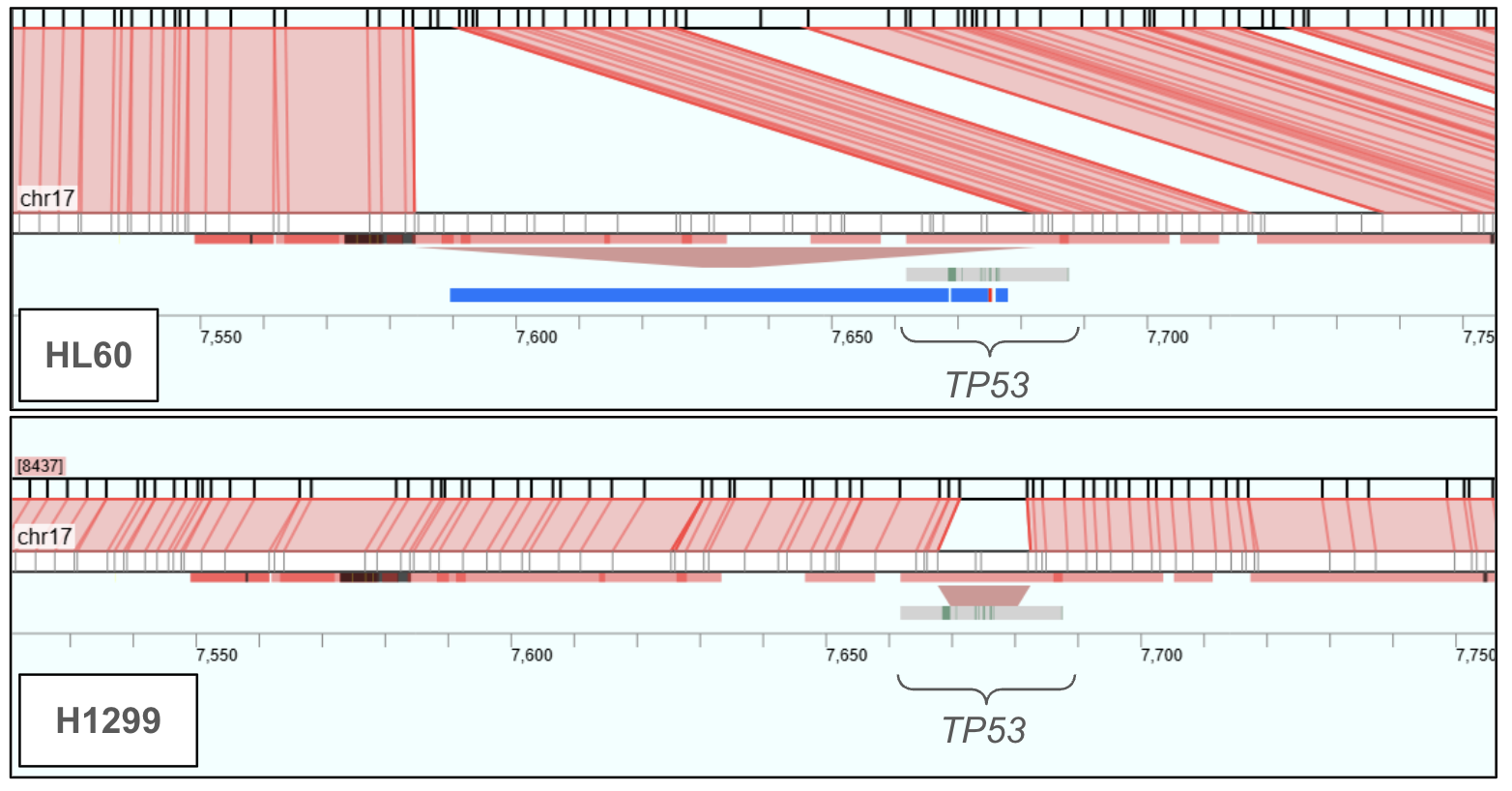
TP53 Deletions
Deletions in the tumor protein p53 (TP53) gene—a critical tumor suppressor—are associated with adverse outcomes across a variety of cancers, including hematologic malignancies. The OhmX Platform identified both large and small homozygous deletions in the TP53 gene.
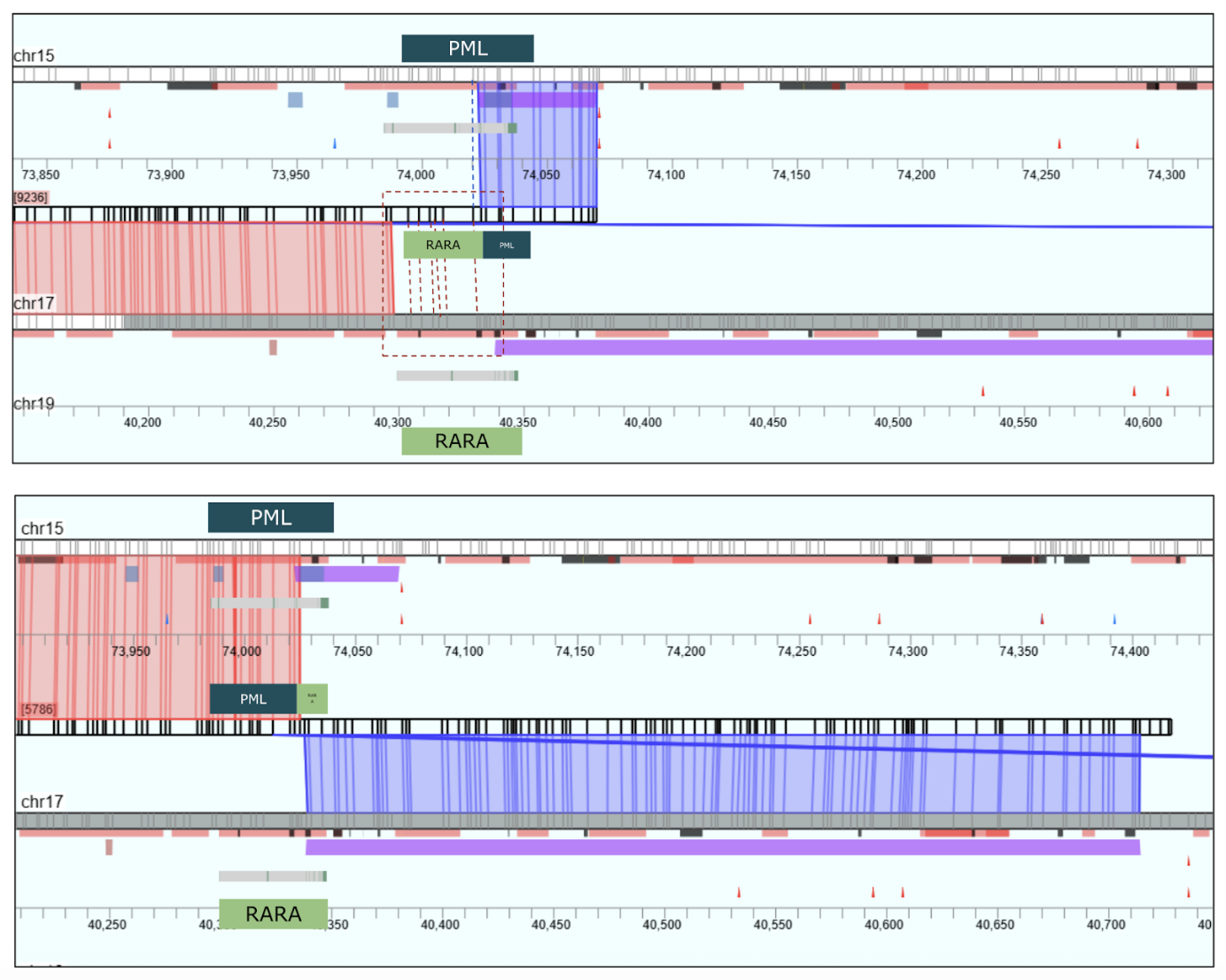
Demonstrating Technical Detection of an Important Gene Fusion (PML::RARA)
The PML::RARA translocation represents a well-known and biologically important gene fusion. In this example, we demonstrate the technical ability of our workflow to detect this variant with precision. The goal is to showcase detection performance — confirming that the platform can resolve a relevant and complex structural event in a research context.
Low-Cost Acquisition Options
Get powerful results without breaking the bank. Our approach delivers exceptional performance at a fraction of the cost compared to alternative approaches.
With zero upfront costs and an 8-genome per month commitment, our Reagent Rental Program makes EGM accessible to all researchers regardless of budget.
Revolutionize Cancer Research with EGM
The OhmX Platform provides the accuracy and clarity you need to tackle the complexities of cancer genomics. Whether you're uncovering unknown SVs or mapping tumor progression, our cutting-edge technology empowers you to obtain actionable insights with confidence.
Our Products
The state-of-the-art OhmX Platform uses electronic nano-detectors to deliver the highest resolution for whole genome structural variant analysis. You can now perform whole genome analysis of SVs down to 300bp in size—enabling insights into previously undetectable DNA variations.
Learn More